The Rise of the Ancients

Forty years ago ‘archaebacteria’ were seen as relatively obscure and mysterious microbes. Now known as archaea, and recognised as a separate domain of life on Earth, they are among the most important and exciting groups of organisms in bioscience, writes Dr Nicholas Robinson
The Biologist 66(5) (RSB 10th Anniversary Special Issue) p16-19
At the end of the 1960s a now outdated five-kingdom system was proposed to classify all forms of cellular life.
It divided organisms into either prokaryotic (or eubacteria, more commonly referred to as bacteria) or the eukaryotic divisions of the fungi, protists, plants and animals.
The name eukaryote is derived from the Greek words eu, meaning ‘true’, and karyon, meaning ‘kernel’ or ‘nut’ in reference to the nucleus – as the name suggests this compartment is absent in prokaryotic cells (‘early’, or ‘prior to’ nucleus). Furthermore, these rudimentary organisms also lack the more complex cellular features associated with the eukaryotes, such as endomembrane systems including the Golgi apparatus, and membrane-bound organelles such as mitochondria and the chloroplasts in plants.
However, having effectively separated all biological life into these taxonomic divisions, the biological community could not have predicted that a whole new category would eventually be required to account for a unique form of life.
In what is arguably one of the most unexpected biological discoveries of the 20th century, a publication at the end of the 1970s astounded the scientific community by proposing a totally new division of life[1], initially referred to as the Archaebacteria and then subsequently renamed as the Archaea (derived from the Greek word archaios meaning ‘archetypal’, ‘primitive’ or ‘ancient’).
These organisms were originally distinguished from other microbes by phylogenetic approaches developed by Carl Woese, George Fox and colleagues, who compared the ribosomal RNA sequences of a large sample of microbes. When Woese sequenced an unusual family of microbes known as the methanogens (anaerobic species that generate methane) he recognised immediately that their ribosomal RNA was completely distinct from that of other microbes. He animatedly noted to his colleagues that these organisms were clearly not bacteria, but instead represented an entirely new form of life.
This conceptual departure was so radical that it was met with great resistance and scepticism by some in the scientific community. However, with time and further investigation, it became clear that biological entities could not simply be divided broadly into two divisions of prokaryotes and eukaryotes. A three-domain categorisation was proposed encompassing the eukaryotes, bacteria and the newly discovered archaea. Reconstructions of the resultant tripartite tree of life suggested that archaea were more closely related to the eukaryotes than the bacteria[2].
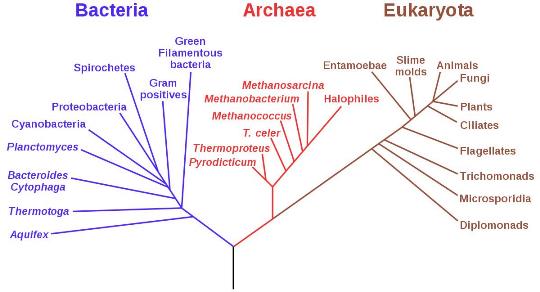 This phylogenic tree, based on the relationship between RNA genes, follows the three-domain system proposed by Carl Wose in 1977. Some later versions of the tree of life place the eukaryotes as a branch within the archaeal domain.
This phylogenic tree, based on the relationship between RNA genes, follows the three-domain system proposed by Carl Wose in 1977. Some later versions of the tree of life place the eukaryotes as a branch within the archaeal domain. Increasing awareness
It is still common to meet individuals outside the immediate scientific communities who are uninformed or even unaware of these organisms. Nevertheless, in just four decades since their discovery, considerable research efforts focused on archaeal biology have been instrumental in advancing our understanding of a number of critical biological processes, some of which have led to important mechanistic insights and biotechnological innovations[3].
Knowledge of the archaea has also improved our understanding of ecosystems associated with a wide variety of important biological niches, with potential far-reaching impact in terms of the environment and world economy[3]. Studies of archaeal biology have provided insights into how life emerged and diversified on Earth, and our understanding of the extremophilic archaea has even helped to redefine the parameters in the search for life beyond this planet[4],[5].
In parallel with an increasing general awareness of the archaea, descriptions of these organisms have more recently appeared in general scientific textbooks and are now also mentioned in blogs, social media sites and general press releases, and consequently an appreciation of these organisms is beginning to enter the collective consciousness of the general population.
Public interest was piqued recently by the discovery of a novel superphylum of archaea, the Asgard archaea[6],[7]. Species within this superphylum are named after the Norse gods, including Loki, Thor, Odin and Heimdall. The first of these organisms to be discovered, the Lokiarchaeota, were found to have genomes encoding for a variety of cellular features that were previously believed to be exclusively eukaryotic innovations.
Soon other related organisms were discovered in different ecological niches around the globe[7]. The Asgardian species genetically encode eukaryotic-like components of the endomembrane systems, small GTPases, membrane-trafficking machineries, vesicle biogenesis proteins, post-translational protein modification systems and cytoskeletal components that are absent in non-Asgardian archaeal cells[7].
 Archaea are generally similar in size and shape to bacteria, but a few forms have intriguing shapes, such as the box-shaped Haloquadratum walsbyi
Archaea are generally similar in size and shape to bacteria, but a few forms have intriguing shapes, such as the box-shaped Haloquadratum walsbyiIndeed, phylogenetic analyses of these organisms suggest an intimate relationship with the eukaryotes, indicating that eukaryotic organisms likely arose from an Asgard-like ancestor or an as-yet-undiscovered sister group[8]. It should be noted, however, that the topology of the tree of life remains an active and intensely contested area of research, hypotheses and discussion. All our understanding of Asgard species is gleaned from bioinformatic interpretation of metagenomic assemblies, as these microbes are yet to be cultured.
Over the last 40 years a growing cohort of dedicated research groups from around the world have advanced our broad understanding of the ecology and metabolism of these fascinating organisms. Investigations of archaeal model systems have also unveiled valuable details regarding the genetics, biochemistry and associated molecular mechanisms of a wide variety of fundamental cellular processes such as cell division, protein homeostasis, DNA replication and genomic repair.
Given the close phylogenetic relationship between the archaea and the eukaryotes, many of these studies have aided our understanding of how the equivalent mechanisms operate in our own cells, and studies of the transcription and translation apparatus from archaea have also proved invaluable in our understanding of these fundamental processes central to all life.
In many cases these studies have taken advantage of the intrinsic biochemical robustness of the proteins and complexes associated with the thermophilic archaea. The ongoing study of archaeal biology will also no doubt lead to discoveries that will unlock future scientific knowledge and economic potential. We take for granted the high-fidelity thermostable polymerases that are utilised in the polymerase chain reaction, and it is clear that the archaea offer a largely unexplored reservoir of other biotechnologically useful enzymes, many of which have significant benefits.
Even the recent development of the high-profile CRISPR-Cas gene editing technology has origins routed firmly in studies of the prokaryotic adaptive immune system in both archaea and bacteria that recognise foreign DNA. CRISPR (clustered regularly interspersed short palindromic repeats) refers to unusual palindromic repeated elements that were first described by Mojica as short, regularly spaced repeats in the halophilic archaea Haloferax mediterranei[9]. Similar repeats had also been observed by Ishino et al[10]. in the bacterium Escherichia coli, and Mojica and others eventually determined that these repeats represented a repository for viral and other foreign DNA elements in an adaptive immune system that protects the host from invading genetic material.
Considerable and incremental research efforts by bioinformaticians, microbiologists and structural biologists have since resulted in a clear understanding of how this system operates and how it could be appropriated as a genome editing tool. (See our interview with Jennifer Doudna, co-inventor of CRISPR gene-editing technology.)
From obscure to everywhere
While the Archaea were for many years often perceived as unusual and obscure organisms, existing exclusively in extreme ecological environments such as the deep-sea hydrothermal vents or hypersaline lakes, it is now known these organisms are considerably more diverse and adaptable, and are not restricted to hostile habitats.
It is now widely accepted that these microbes have occupied almost every imaginable ecological niche in the biosphere, and contribute critically to biomass and global nutrient cycling in our oceans, soils and other ecologically important environments[11],[12].
For example, archaea have recently been shown to make important contributions to ammonia oxidation, the first step of nitrification in the nitrogen cycle[13] and Archaea are solely responsible for all the world’s biological generation of methane, a critical greenhouse gas. It is also becoming clear that the presence of important archaea in microbial communities has been significantly underrepresented to date owing to the insensitivity or lack of specificity of previous detection methods.
Furthermore, there is increasing recognition of the importance of archaea in the microbiomes of both plants and animals[14], and with advanced detection methods archaeal populations have been identified in a range of microbiomes including plant root systems, and human skin, nasal cavities, lungs and the digestive system[14].
To date, archaea have not been found to be the causative agent of any plant or animal disease, but it is clear that interactions and transactions between archaeal and bacterial microbes within a community may have critical biological ramifications, modulating or potentiating the virulence of an infection or altering the availability of nutrients in the rhizosphere of a plant[15].
It therefore seems probable that the study of archaeal populations within microbiomes will have repercussions in important areas such as food sustainability and health. In addition, it is worth considering that archaeal species are also likely to harbour unexplored reservoirs of novel antibiotic molecules and systems.
As we enter the fifth decade of biological investigation of archaeal organisms these widespread but often overlooked microbes have provided new avenues of biological study, improving our understanding of ecosystems, microbiomes and fundamental cellular mechanisms.
With the increasing awareness of the existence and biological importance of the archaea, the number of researchers studying archaeal biology is steadily increasing and ongoing and future study of this broad division of life offers exciting potential for discoveries.The spotlight on archaeal biology has never been brighter and this field offers great opportunities for the next generation of researchers.
- This article is an edited version of an editorial that appeared in a special issue of Emerging Topics in Life Sciences, a journal co-owned by the RSB and the Biochemical Society. The issue (Vol 2 Issue 4) provides a snapshot of the latest developments in archaeal biology.
1) Woese, C. R. & Fox, G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 74 (11) 5088– 5090 (1977).
2) Eme, L. & Doolittle, W. F. Archaea. Curr. Biol. 25, R851–R855 (2015).
3) Jarrell, K. F. et al. Major players on the microbial stage: why archaea are important. Microbiology 157, 919–936 (2011).
4) Cavicchioli, R. Extremophiles and the search for extraterrestrial life. Astrobiology 2, 281–292 (2002).
5) Schulze-Makuch, D. et al. The adaptability of life on earth and the diversity of planetary habitats. Front. Microbiol. 8 (2017).
6) Spang, A. et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015).
7) Zaremba-Niedzwiedzka, K. et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017).
8) Eme, L. et al. Archaea and the origin of eukaryotes. Nat. Rev. Microbiol. 15, 711–723 (2017).
9) Mojica, F. J. et al. Long stretches of short tandem repeats are present in the largest replicons of the archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 17, 85–93 (1995).
10) Ishino, Y. et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433 (1987).
11) Adam, P. S. et al. The growing tree of archaea: new perspectives on their diversity, evolution and ecology. ISME J. 11, 2407–2425 (2017).
12) Spang, A. et al. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357, eaaf3883 (2017).
13) Offre, P. et al. Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67, 437–457 (2013).
14) Moissl-Eichinger, C. et al. Archaea are interactive components of complex microbiomes. Trends Microbiol. 26, 70–85 (2018).
15) Braga, R. M. et al. Microbial interactions: ecology in a molecular perspective. Braz. J. Microbiol. 47(1) 86–98 (2016).


