Focus on: Computer-aided biology

Will augmenting bioscience with computer-aided design software change what it means to be a biologist?
The Biologist 66(3) p30-31
In the 1970s and 1980s the design of silicon circuits became so complex that creating blueprints for them graphically started to become impossible.
A range of software tools was developed to help engineers create circuits using programming language, vastly simplifying the design process and increasing the complexity of circuits that could be built.
As these tools became more sophisticated, computing power was used to test the performance and optimise the designs of experimental circuits, reducing the need to actually manufacture physical prototypes.
Now, in all areas of engineering that involve very complex systems, computer-aided design (CAD) is used in the creation, modification, analysis and optimisation of designs.
Bioengineers and synthetic biologists are increasingly taking a similar approach, developing tools that design cellular pathways and genetic circuits for biologists. In what is known as computer-aided biology (CAB) or Bio-CAD, a biologist enters the desired output – for example, the production of a particular protein under certain conditions – and the software crunches data to calculate a DNA sequence that will make this happen in the most efficient or effective way in a particular organism.
Christopher Voigt, a professor of advanced biotechnology at MIT and developer of the Bio-CAD tool Cello, says designing genetic circuits with machine learning tools often creates surprising metabolic pathways that are not found in nature and would be unlikely to be intuited by a biologist.
“One of the interesting things is the more powerful these tools, the less and less the design is like how biology does it,” says Voigt. The difference between using ‘evolved’ biological systems and computer-designed ones can be huge, he says.
Dr Guy-Bart Stan, head of the Control Engineering Synthetic Biology Group at Imperial College London, says that the way most biologists introduce new gene functions into organisms remains, to some extent, trial and error. “You need to work out the strength of certain promoters, determine ribosome binding sites and codon modifications for the protein sequence... biologists have, until now, been brute-forcing the problem – growing many different strains of organisms and using high-throughput screening to select the best ones,” he explains.
This approach has undoubtedly led to many advances and successful products, but he hopes the development and wider use of Bio-CAD can make bioscience more systematic and rigorous. “Any other industry that works with very complex systems uses this approach – and biotechnology should be no different,” says Stan.
Although there is a growing number of Bio-CAD tools in development – Benchling, Cello, J5 and SynBioHub being some of the best known – the number of bioscientists using these tools remains small, possibly because the software’s interfaces and programming languages are obscure to all but those from engineering or computer programming backgrounds.
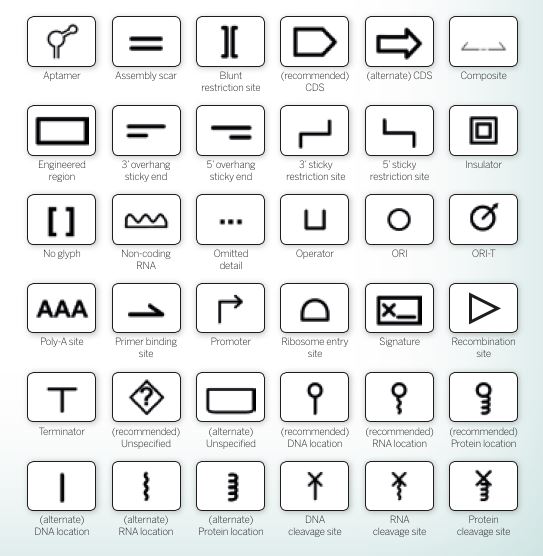 A small selection of glyphs used to represent components of genetic circuits in SBOL, the synthetic biology open language
A small selection of glyphs used to represent components of genetic circuits in SBOL, the synthetic biology open languageAs tools are developed that focus on different aspects of biologists’ workflow, there is a need for a standard language and file formats that enable various tools to talk to each other. This looks likely to be based on SBOL, the synthetic biology open language currently used as a global standard for the exchange of synthetic biology data and designs. As part of this, a standard series of glyphs (symbols) has emerged to visually represent genetic and biochemical circuits (see above).
Developments in Bio-CAD could eventually lead to entirely automated synthetic biology workflows, with optimal biochemistry and genetic circuits calculated by computer, and the actual assembly and cultivation of the modified organisms controlled by liquid-handling robots. “That is the dream,” says Stan. “There will always be a need for specific interventions in the laboratory that cannot be automated fully.
But what you want is a paradigm shift: the human effort should be displaced from the low-level, routine cultivation and repeated experiments in the laboratory to the high-level design ideas for the problem and optimisation of its solutions.”
- Look out for the forthcoming synthetic biology special issue of the RSB's journal Emerging Topics in Life Science, published in partnership with the Biochemical Society.

BIOLOGY IN BINARY Computer-aided biology (CAB) is the term given to a suite of tools that augment human capabilities in biological research.
It can include software for simulating and designing biological systems, as well as systems that can design, simulate, optimise and execute experiments.
Many research teams and companies are working towards an integrated suite of tools that will be comparable to CAD-based workflows used in other sectors such as the aerospace, automobile or telecommunications industries.
Bioengineers believe moving towards this fully digital-to-automated workflow will vastly reduce the cost and time to market for drugs and other biological products.


