"The wave has only got bigger"

Co-inventor of CRISPR gene-editing technology Jennifer Doudna talks to Tom Ireland about one of the biggest science stories of the decade
The Biologist 66(5) (RSB 10th Anniversary Special Issue) p10-15
In 2012 American biochemist Jennifer Doudna and her collaborator, Emmanuelle Charpentier, proposed that CRISPR – a group of genes and molecules used by bacteria to recognise and destroy viral DNA – could be re-purposed as a powerfully simple and programmable gene-editing tool.
Their work led to an explosion of interest in CRISPR and exciting new applications of gene-editing across all areas of biology. With its unprecedented efficiency and ease of use, CRISPR has not only supplanted all previous genetic engineering technologies, but has revolutionised what is possible in life science – at the same time raising profound questions about what society should and should not do with such powerful technology.
Born in Washington DC, Doudna moved to Hilo, a large town in Hawaii, aged seven. After briefly considering ditching chemistry to major in French at college, she stuck with science, eventually gaining her PhD in biological chemistry and molecular pharmacology from Harvard in 1989.
Before CRISPR, Doudna and her laboratory (now based at the University of California, Berkeley) focused on how RNA molecules can act as chemical catalysts as well as messengers in cells. Doudna’s laboratory was the first to deduce the structure of a ribozyme – that is, a length of DNA that acts like an enzyme.
Doudna then focused on RNA interference, a process in which RNA molecules regulate gene expression or translation. It was this work that led to a meeting with Jill Banfield, a Berkeley microbiologist who at the time was trying to understand mysterious sequences of bacterial DNA known as clustered regularly interspaced short palindromic repeats – or CRISPR for short.
After years at the centre of intense media interest and patent disputes over the gene-editing technology, Doudna continues to work on the fundamental biology of CRISPR bacterial defence systems, of which there are many types, and she is a key figure in discussions on the safety and ethics of CRISPR-based technologies.
TI: So many interesting breakthroughs in science involve chance conversations, introductions and collaborations – tell me about why Jill Banfield introduced you to these weird bacterial DNA sequences called CRISPR when you were working on RNA interference in eukaryotic cells.
JD: Well, it was exactly because we were working on RNA interference that my laboratory and our research came to Jill’s attention. She was studying bacterial genomes, and particularly these DNA sequences that, at the time, were of unknown function, but had signatures that made them appear to be made up of viral DNA.
Jill and just a handful of others at the time had postulated that these curious sequences, which had come to be called CRISPR, might be some kind of RNA-based adaptive immune system, although nobody at the time had any experimental evidence of that. Crucially Jill had the insight that these systems might be controlled by RNA, and that was her purpose in reaching out to me.
It sounded possible that bacteria had evolved a system for defending against viruses that was in some ways very similar to the RNA interference that goes on in eukaryotic cells.
 Jennifer Doudna in her lab at UC Berkeley.
Jennifer Doudna in her lab at UC Berkeley. Do you remember your first introduction to the intricacies of CRISPR, which was of course completely obscure and unknown at the time?
Yes, I remember it very well. Jill and I met at a coffee bar on the Berkeley campus known as the Free Speech Movement Café – a very Berkeley sounding place!
We were sitting out on the patio and Jill had a big stack of papers in front of her. She was showing me all these examples of repetitive DNA sequences from bacteria that seemed to include sequences that came from viruses. At the time, for me, it was like drinking from a fire hose. There was a lot of data and I was struggling to understand, but Jill was clearly very excited and I really had a feeling that this was something potentially very interesting.
I’ve always had a fascination with the underlying theme of evolution in biology, and how a lot of biological pathways, systems and organisms are connected by evolution. You can find patterns of similarity even in seemingly very different kinds of cells or molecular pathways. So when I heard about the possibility of an RNA-guided system in bacteria, I was immediately interested.
When did you start making the connection that this intriguing bacterial immune system could be used to target and cut particular sequences of DNA?
The first three or four years of CRISPR biology in our laboratory was focused on Cas proteins that recognise CRISPR RNA and cut it up into useful, usable segments of RNA. It was very similar to work we had been doing on RNA interference and an enzyme that chops up RNA called Dicer.
A graduate student in my lab, Rachel Haurwitz, who had been taking some classes at Berkeley’s Business School, got me thinking about starting a biotechnology company. Rachel and I founded a company called Caribou Biosciences in October 2011 with the purpose of developing these CRISPR-Cas proteins for research or clinical uses – for example, as diagnostic tools that could recognise and signal the presence of viral RNA molecules.
This was before our laboratory had completed our work on the class of proteins known as Cas9. Of course, once the CRISPR-Cas9 work was done it was clear this was going to be a very useful way to change DNA sequences and therefore genomes – and that changed lots of things. It certainly changed the goals of Caribou at that point.
Was there a single ‘eureka’ moment when the power of the CRISPR-Cas9 system dawned on you? Or was it more stepwise and iterative than that?
Martin Jinek, who was a postdoc in the laboratory doing the research on Cas9, did the initial experiments on Cas9. He showed it was programmable – an RNA-programmed protein – and figured out how the DNA targeting and cutting works. Then, very importantly, he figured out that he could simplify the system, relative to the way it works in nature, combining two types of RNA into one RNA to guide Cas9 to its target.
When he showed that approach worked very nicely for directing Cas9 to a desired sequence for cutting, that was really when we said “Gosh – this is going to be amazing”. It was just so easy to work with this protein – a lot easier than any of the earlier technologies for gene engineering.
In your book, A Crack in Creation, you wrote of having a recurrent dream that a tsunami is approaching you on a beach near where you grew up, representing the overwhelming surge in interest and questions raised by this technology. Do you still have this feeling?
Absolutely. The wave has only got bigger.So many things are happening right now on multiple fronts and it really cuts across every area of biology. I find that both very, very exciting, and also a bit daunting.
It’s difficult to keep up with the scientific literature, much less stay on top of all the different facets of genome editing such as questions about ethics, safety and the affordability of future gene- editing therapies.
 Jennifer Doudna in the lab using a combination of cryo-electron microscopy and 3D image reconstruction
Jennifer Doudna in the lab using a combination of cryo-electron microscopy and 3D image reconstructionGoing back to that period when things really exploded, around four or five years ago, how did you feel being at the heart of so many concerns and difficult ethical questions everyone suddenly wanted answers to?
I was incredibly nervous. I felt poorly equipped to answer any of those concerns and the really big ethical questions. I was hesitant, initially, to dive into any kind of public discussion about these issues, and it was a very intense time for me.
Thinking about how genome editing might eventually be deployed, especially in places like the human germline, quickly moved from being science fiction to becoming clearly on the scientific horizon. I started to think very seriously, first of all, about how we alert people to what is happening in this area of science. Then, how would one start to engage with all of the different kinds of stakeholders that could play a role in making decisions around a technology such as this? I felt a real sense of personal responsibility having been involved in the early stages of this whole field.
I spent a lot of time in 2014 talking with colleagues of mine here at Berkeley and at the Innovative Genomics Institute, which I started that year. In January 2015 we convened the first meeting on human genome editing in the Napa Valley. That was really the kick-off to those sorts of conversations, because after that the National Academies and the international scientific and clinical organisations got involved.
Since then we’ve just seen an incredible uptick in the numbers of meetings, reports and groups that are weighing in on what should be done now that we have access to such a powerful tool.
How do you feel that those conversations are going?
On the whole I would say that I’m quite pleased. I’m first of all glad that they’re happening at all – I think that’s paramount. It’s also important that they be made open to people who are not specialists in the field, or maybe don’t even have particular scientific training.
There’s been a real appreciation across the scientific community that it’s critical for scientists to be involved in [these ethical] discussions, even though for many scientists it’s just not in our DNA – a lot of us would prefer to be in the laboratory doing the next experiment and not talking on a public platform about something that could be quite contentious.
Obviously some of the most contentious debate around gene editing involves the human germline. Do you think things are moving too quickly here?
Yes, I do, and I don’t think I am alone in having that view. The data that was presented by He Jiankui last year [the Chinese scientist who claimed to have used CRISPR to confer HIV resistance to twin girls] demonstrated two things very clearly.
First, that the technology is just not ready for use in human embryos, not for any kind of clinical application. We don’t have a good enough handle on how to control DNA repair to ensure that accurate changes are made to the genomes. That’s on the technical side.
Perhaps more importantly it was clear that there was no good plan in place for either consent in the patients who were involved in that study or in following up with the babies after birth to ensure that their health was monitored.
 Hilo, on Hawaii's Big Island.
Hilo, on Hawaii's Big Island. In your book you said it was “inevitable” that someone was going to use CRISPR to change the human germline. Is that because the public will eventually demand it or because you think there will always be some scientist or state that does it regardless of regulations?
It was clear this would be something that people would find attractive and something that deserved exploration. You could, in principle, remove genes from a family line that cause genetic disease, for example.
There’s inevitably a lot of fascination with the potential to change the human germline – it’s a very powerful thing to think about. It also opens the door to engineering humans or changing the course of human evolution if you took it far enough.
Then I think there’s an aspect of defiance, which involves fame and public notoriety, that is not unique to CRISPR. It’s true for any area of science and technology where there’s a very obvious opportunity to do something that is a first and will attract a lot of public attention. This certainly fell in that bucket. I think for those reasons it just seemed to me that the use of CRISPR in human embryos was inevitable.
What is your view on those who say that tools such as CRISPR should be made available to non-scientists to use in non-institutional settings – for example, the US biohackers who have claimed to have injected themselves with CRISPR or are trying to develop DIY gene therapies?
Part of the whole biohacker movement is really interesting because it opens up science to many more people than just those who have PhDs, and in principle that’s appealing to me. I like the idea that science becomes more of a community effort and something of societal interest, where people have ways of gaining access to tools and technologies like this.
That said, I don’t think it’s a great idea to inject anything into yourself, much less something that could cut or damage your DNA. So I don’t recommend it.
Can you tell me a bit about what the Doudna laboratory is working on at the moment?
We are continuing to explore the fundamental biology and functions of CRISPR systems in many different kinds of microbes. There is a lot of variability in how these systems are put together.
My laboratory, like many others, is also very enthusiastic about how we could use genome editing, especially the opportunities to uncover the cause of a disease and ultimately find treatments or even cures for genetic disease.
We are very focused on how to deliver gene-editing molecules into cells and tissues, as well as how to understand the outcomes of gene editing, ensuring editing is done correctly and that edited genes don’t wreak unexpected changes themselves.
Do you have a favourite application of gene editing that just strikes you as really neat or interesting?
There are so many. One of the recent ones I’ve been talking about is from developmental biology. We know that snails’ shells curl in a right-hand direction – you very rarely find a left-handed, left-curling snail in nature. Developmental biologists have been trying to understand the genetics of this for a long time.
With CRISPR it was possible to figure out not only the genes that were part of this handedness, but also to ultimately identify one single gene that when knocked out actually created left-handed snails. It’s a question that’s been around forever and finally it’s possible to answer it – not in a model system or by inferring behaviour from cells, but from making changes in the organism itself.
Another thing worth mentioning is the use of CRISPR with a mechanism called gene drive, where traits are introduced that spread very quickly throughout populations – this could, in principle, prevent mosquitoes from being able to spread diseases such as dengue or zika virus, and malaria. Of course, this comes with significant environmental risk, but I think it is being pursued with appropriate caution.
Right now, the data looks very good and it’s moving forward very quickly. The biggest global impact, at least in the near term, will likely be in agriculture, because of the potential to alter plant traits that will make plants more resistant to climate change and to pests, more nutritious or even to develop in new environmental niches. That’s fascinating to me.
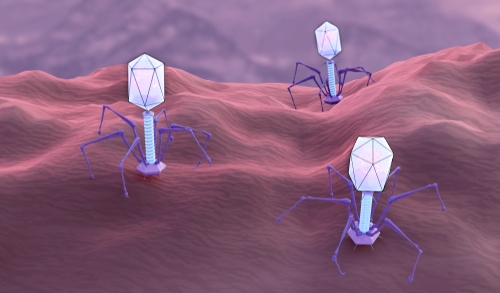 CRISPR-Cas9, now famous for its use as a gene-editing tool, is one of many CRISPR systems that evolved in bacteria and archaea as defences against phages. Viral DNA sequences are captured and integrated into the CRISPR region of the host’s genome, forming the basis of an adaptive immune system that can then silence or destroy DNA containing those sequences in future. Phages are thought to be the most abundant unit of life on Earth, with some environments containing 10 times as many phages as bacterial cells. The war between these viruses and prokaryotic cells – “the oldest war on Earth”, according to Doudna – has driven the evolution of a range of complex molecular systems that target viral DNA, as well as viral counter-defences, including ones that can shut down CRISPR.
CRISPR-Cas9, now famous for its use as a gene-editing tool, is one of many CRISPR systems that evolved in bacteria and archaea as defences against phages. Viral DNA sequences are captured and integrated into the CRISPR region of the host’s genome, forming the basis of an adaptive immune system that can then silence or destroy DNA containing those sequences in future. Phages are thought to be the most abundant unit of life on Earth, with some environments containing 10 times as many phages as bacterial cells. The war between these viruses and prokaryotic cells – “the oldest war on Earth”, according to Doudna – has driven the evolution of a range of complex molecular systems that target viral DNA, as well as viral counter-defences, including ones that can shut down CRISPR.
Jennifer Doudna is the Li Ka Shing Chancellor’s Chair and a Professor in the Departments of Chemistry and of Molecular & Cell Biology at the University of California, Berkeley, as well as an Investigator at the Howard Hughes Medical Institute.


