All together now
Could a diverse group of compounds known as oxylipins, found across the kingdoms of life, be the molecules that enabled ancient single cells to join forces and become multicellular? Georgy Kurakin and colleagues look at the evidence
October 6th 2021
Not everybody has heard of oxylipins, but everyone will have experienced their effect in the body. And everybody who has ever taken aspirin, ibuprofen or an anti-asthmatic drug has blocked their effect. These derivatives of fatty acids are among the first molecules produced in reaction to injury. When something disrupts our cells — whether that be a toxic streptococcus, an invading coronavirus or a hot iron, the cells begin synthesizing and releasing oxylipins. One could say that oxylipins are our cells’ cry for help: the signals that help kick-start inflammation, the nonspecific reaction of our tissues to injury, causing leukocytes and thrombocytes to come to the rescue through dilated capillaries.
The oxylipins found as signalling molecules in animals are known as eicosanoids, associated with a vast range of functions and potential functions beyond inflammation and cell-signalling. But oxylipins are found across the kingdoms of life.
Plants, lacking mobile immune cells flowing round a bloodstream, have a very different way of responding to injury, producing a wide array of bactericidal, fungicidal and insecticidal compounds in response to injury. But the process also begins with oxylipins being synthesised from fatty acids[1]. While our bodies use local-acting oxylipins like prostaglandins and leukotrienes to trigger inflammation, plants produce jasmonates, a class of plant hormones that coordinate not only wounding responses, but also other stress reactions like reaction to drought and excessive salinity.
Some plant oxylipins are volatile compounds that can diffuse from plant to plant by air, warning neighbouring plants about danger and helping them elicit defence responses in advance. The pleasant smell of freshly cut grass contains these volatile oxylipins — the chemical scream of plant tissues injured en masse. Oxylipins have been found to trigger immune responses in red algae and even diatoms[2,3,4], and oxylipin synthesis as a result of stress has been described in corals[5,6].
Some eukaryotes use oxylipins differently: in animals, they may influence cell function and differentiation and some may even be anti-inflammatory; brown algae use them as pheromones, allowing male and female gametes to find each other[7]; fungi use them to signal to their neighbours that space is limited, which can prompt neighbours to switch to a more compact form (i.e. from mycelium to yeast)[8,9], and fungi also use oxylipins to induce their conspecifics to reproduce sexually[8]. When comparing even completely dissimilar multicellular eukaryotes, their cells appear to use oxylipins in their chemical ‘conversations’ between cells and between individuals. So, we asked, what is the evolutionary origin of this biochemical Esperanto?
 Oxylipins are key signalling molecules in the initiation of inflammation in animals (pictured) and wound responses in plants. Image via the Department of Pathology at Calicut Medical College.
Oxylipins are key signalling molecules in the initiation of inflammation in animals (pictured) and wound responses in plants. Image via the Department of Pathology at Calicut Medical College.There is scarce experimental data on the function of oxylipins in bacteria and simple eukaryotes like protozoa. We know that some bacteria can synthesise oxylipins, but do not know what they do it for. We performed bioinformatic analysis of large biological databases to shed some light on the question, with the results published recently in Biochemistry (Moscow)[10].
Lipoxygenase enzymes (also known as LOX enzymes) are a convenient peg to hang the research on. This protein participates in oxylipin biosynthesis in all organisms we have discussed and is very conserved — its structure and function are very similar even in distantly related organisms, from Irish moss to the house mouse. It is also instantly recognisable when analysing protein or nucleic acid sequences. So, this enzyme is a fair ‘mark’ to track the evolution of oxylipin signalling.
A previous bioinformatic study had shown only a small number of bacteria have lipoxygenases — approximately one in 200 species[11]. Our more detailed statistical analysis confirmed that lipoxygenases are very rare in most bacterial (and protozoan) taxa, but in a few taxa they are exceptionally abundant. Interestingly, these taxa have a significant distinction: they form simple multicellular structures.
 Cyanobacteria, shown here forming long chains of cells, are thought to be the ‘inventors’ of multicellularity – and may be the inventors of lipoxygenases, too. Image courtesy of Josef Reischig’s archive via Wikimedia Commons.
Cyanobacteria, shown here forming long chains of cells, are thought to be the ‘inventors’ of multicellularity – and may be the inventors of lipoxygenases, too. Image courtesy of Josef Reischig’s archive via Wikimedia Commons.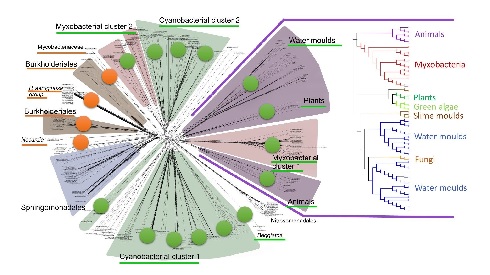 (Click image to enlarge) The author’s bioinformatic data on lipoxygenase evolution depicted in one figure. Due to multiple horizontal gene transfers, their evolution is more net-like than tree-like, with lipoxygenases associated with multicellular and multicellular-like groups (green marks) and are also linked to some virulent groups (orange marks). Image courtesy: Kurakin, G.F. (2020) Biochemistry (Moscow).
(Click image to enlarge) The author’s bioinformatic data on lipoxygenase evolution depicted in one figure. Due to multiple horizontal gene transfers, their evolution is more net-like than tree-like, with lipoxygenases associated with multicellular and multicellular-like groups (green marks) and are also linked to some virulent groups (orange marks). Image courtesy: Kurakin, G.F. (2020) Biochemistry (Moscow).Cyanobacteria were also the first inventors of multicellularity on our planet — providing another strong evolutionary link between lipoxygenases and multicellularity. It is worth noting that not all organisms require lipoxygenases (and therefore oxylipins) to be multicellular. Even in invertebrates, which contribute more than 95% to all multicellular organisms on Earth, the percentage of lipoxygenase-containing species is lower than 1%. Clearly, multicellular life is possible without LOX.
It’s also worth noting that a few groups of bacteria acquired lipoxygenases but remained single-celled. Our results suggest these organisms could be using them as virulence factors (molecules that enable a pathogen to infect a host and cause disease). This way of using lipoxygenases has been experimentally characterized for Pseudomonas aeruginosa[13], and our results suggest this may also be the case in many other dangerous opportunistic pathogens, such as Pantoea ananas, Burkholderia gladioli, and Pluralibacter gergoviae. Such a mechanism had been earlier proposed for some other pathogens, like the protozoan Toxoplasma gondii, notorious for its ability to cause latent infections in human beings and animals[14]. Maybe LOXs are used for virulence by even more pathogens that we assume.
We have hypothesized that bacterial lipoxygenases generate false ‘attack over’ signals that suppress the immune responses of their hosts and we plan to do a more detailed phylogenetic study of this group to find more evidence or this mechanism. Such studies may lead to the new insights that could aid the development of urgently-needed new antibiotics.
Despite being interesting, the pathogenic function of lipoxygenases (and oxylipins) seems to be secondary to multicellularity (even in most primitive forms observed in bacteria and protozoa) as the primary function. Presumably, ancient bacteria and protozoa used oxylipins like we do — for cell-to-cell signaling. A small amount of experimental data is available to support this — for example wounded cells of the cyanobacterium Nostoc punctiforme produce more oxylipins than non-wounded cells[15], the myxobacterium Myxococcus xanthus twitches more actively sensing oxylipins[16], and the slime mould Dictyostelium discoideum cannot form a normal fruiting body if its lipoxygenase is muted[17].
 Fruiting bodies of Dictyostelium discoideum at various stages of development. The slime mould cannot form fruiting bodies if its lipoxygenase is muted. Image courtesy of Jeff Smith.
Fruiting bodies of Dictyostelium discoideum at various stages of development. The slime mould cannot form fruiting bodies if its lipoxygenase is muted. Image courtesy of Jeff Smith.Learning the language
We do not know yet what bacterial and protozoan cells say to each other in this oxylipin ‘Esperanto’ — but we are almost sure they do say something. Our results also raise an intriguing question: what was the role of oxylipins in the emergences of multicellularity? Clearly, multicellular life is possible without lipoxygenase and oxylipins, but they may have played a pivotal role in its origin.
The path is reminiscent of the history of programmed cell death described by Nick Lane in his well-known story Origins of Death[18]. Programmed cell death (or apoptosis) is necessary for cell differentiation and development in complex multicellular organisms. Tracking its evolutionary history through conserved proteins leads to very similar assumptions: the process seems to have originated in cyanobacteria and was borrowed by the ancestors of multicellular organisms many times. So, bacteria have taught our cells to die. Our research suggests that they could also have given our cells a useful way to communicate, too.
We now plan to follow up our research and to track oxylipin evolution through other enzymes — for example, homologs of cyclooxygenase. However, these methods are unable to tell us what cyanobacterial or slime mold cells actually ‘say’ to each other in this chemical language. We hope that our data could help to develop new experimental strategies of searching oxylipin functions in a more targeted manner.
The authors acknowledge Alina Lebedeva (Tver State University, Faculty of Foreign Languages and International Communication, Teaching Assistant at the Department of Language Theory, Translation and French Philology) for literary editing this article.
1. Heldt, H.-W., Piechulla, B. Lipids are membrane constituents and function as carbon stores. Plant Biochemistry (pp. 359–398), Elsevier (2011)
2. Bouarab, K. et al. The innate immunity of a marine red alga involves oxylipins from both the eicosanoid and octadecanoid pathways. Plant Physiology, 135(3), 1838-1848 (2004).
3. Paul, C., Pohnert, G. Induction of Protease Release of the Resistant Diatom Chaetoceros didymus in Response to Lytic Enzymes from an Algicidal Bacterium. PLoS ONE. 8 (2013).
4. Meyer, N. et al. Algal Oxylipins Mediate the Resistance of Diatoms against Algicidal Bacteria. Marine Drugs, 16, 486 (2018)
5. Lõhelaid, H. et al. Lipoxygenase-allene oxide synthase pathway in octocoral thermal stress response. Coral Reefs, 34, 143–154 (2015).
6. Lõhelaid, H. et al. Up-Regulated Expression of AOS-LOXa and Increased Eicosanoid Synthesis in Response to Coral Wounding. PLOS ONE, 9(2)(2014).
7. Rui, F., Boland, W. Algal pheromone biosynthesis: stereochemical analysis and mechanistic implications in gametes of Ectocarpus siliculosus. The Journal of organic chemistry, 75(12), 3958-3964 (2010).
8. Brodhun, F., Feussner, I. Oxylipins in fungi. FEBS Journal, 278, 1047-1063 (2011).
9. Horowitz Brown, S. et al. Morphological Transitions Governed by Density Dependence and Lipoxygenase Activity in Aspergillus flavus. AEM, 74, 5674-5685 (2008).
10. Kurakin, G.F. et al. Bacterial and Protozoan Lipoxygenases Could be Involved in Cell-to-Cell Signaling and Immune Response Suppression. Biochemistry (Moscow), 85(9), 1048-1063 (2020).
11. Horn, T. et al. Evolutionary aspects of lipoxygenases and genetic diversity of human leukotriene signaling. Progress in Lipid Research, 57, 13-39 (2015).
12. Porta, H., Rocha-Sosa, M. Lipoxygenase in bacteria: a horizontal transfer event? Microbiology (Reading), 147(Pt 12), 3199-200.
13. Morello, E. et al. Pseudomonas aeruginosa lipoxygenase LoxA contributes to lung infection by altering the host immune lipid signaling. Frontiers in microbiology, 10, 1826 (2019).
14. Aliberti, J., Bafica, A. Anti-inflammatory pathways as a host evasion mechanism for pathogens. Prostaglandins, Leukotrienes and Essential Fatty Acids, 73(3-4), 283-288.
15. Lang, I., Feussner, I. Oxylipin formation in Nostoc punctiforme (PCC73102). Phytochemistry, 68(8), 1120-1127.
16. Jung-Ung An, Deok-Kun Oh. Stabilization and improved activity of arachidonate 11S-lipoxygenase from proteobacterium Myxococcus xanthus. Journal of Lipid Research. 59, 2153-2163 (2007).
17. Phenotype and Strain Details for LipA-. DictyBase.
18. Lane, N. Marine microbiology: origins of death. Nature News, 453(7195), 583-585 (2008).
Georgy Kurakin AMRSB is a teaching assistant at the Faculty of Biomedicine of Pirogov Russian National Research Medical University (Russia). His research interests include evolutionary biology and bioinformatics and he has contributed to Russian popular science magazines and the British magazine Laboratory News.
Anna Samoukina PhD is the Head of the Scientific Department of Tver State Medical University (Russia) and the Head of the Department of Hygiene and Ecology of the same university. She studies human microbiome and its influence on human health and diseases.
Nadezhda Potapova is a Research Associate of Institute for Information Transmission Problems of the Russian Academy of Sciences (Kharkevich Institute). Her area of expertise is evolutionary genomics.


