Accelerating Evolution

Artificial gene drives are a way of spreading genetic changes through a population quickly – for example, to make large numbers of wild mosquitos infertile or unable to carry malaria. Bruce Whitelaw and Gus McFarlane explain how this technology works, and how it could be used in conservation and agriculture as well as public health
The Biologist 66(6) p18-21
It was the work of Gregor Mendel and Charles Darwin that revealed the fundamental mechanisms of genetic inheritance. Mendel observed consistent patterns of inheritance from parents to offspring and Darwin’s theory of natural selection illustrated how traits are selected over time based on fitness.
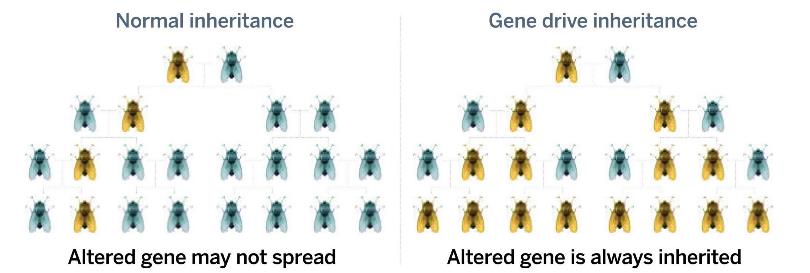
However, we now know inheritance does not always follow the rules. In some cases the odds of a particular gene being transmitted to the next generation can be heavily skewed. Genetic elements that manage to beat the odds by being passed on more than half the time are known as ‘gene drives’, and this numerical advantage enables them to spread through populations even if they reduce an individual animal or plant’s fitness.
Gene drives evolve in sexually reproducing species due to the genetic conflict that occurs between genes in an organism, enabling them to gain an advantage over their genetic competition and increase their likelihood of being passed to future generations. While most genes are transmitted to 50% of the offspring, gene drives manage to be transmitted to more than 50% of the offspring – that could be just 50.01% or a full 100%.
Broadly speaking there are two main strategies genes can use to achieve ‘drive’. The first is interference, whereby the gene gains an advantage by disrupting the transmission of an alternative gene. For example, a gene in a diploid organism could reduce the fitness of the 50% of sperm or eggs that receive the alternative gene during the process of gametogenesis, thereby increasing its own chance of success and reproduction in the next generation.
The second method is by over-replication. These gene drives bias their transmission to the next generation by being replicated more than other genes in an animal. Homing endonuclease genes are a good example: these cut DNA and get copied into the cut site as part of the repair process, increasing their abundance in the animal’s genome[1].
Learning to drive
It was the study of naturally occurring gene drives that lead Austin Burt of Imperial College London to first propose that synthetic gene drives could be used to alter or suppress wild animal populations[2].
That was over a decade ago, and the genome editing tools available at the time were not useful in many organisms or suffered from evolutionarily instability. Fast forward to 2019 and the blockbuster genome editing tool known as CRISPR has enabled scientists to engineer synthetic gene drives in animals that could profoundly benefit public health, agriculture and the environment.
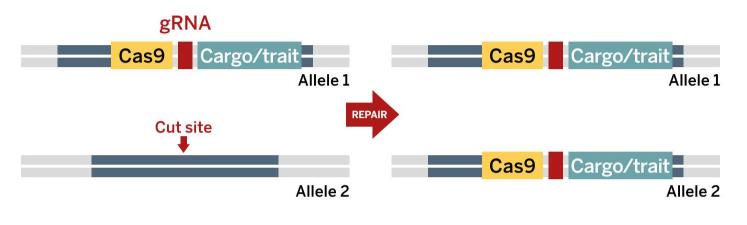 A ‘gene drive cassette’ is inserted into an organism’s genome on one chromosome of a homologous pair. When the Cas9 and guide RNA are expressed, together they make a cut at the same location on the other chromosome of the pair.
A ‘gene drive cassette’ is inserted into an organism’s genome on one chromosome of a homologous pair. When the Cas9 and guide RNA are expressed, together they make a cut at the same location on the other chromosome of the pair.The cell repairs the cut by copying the sequence at this location from the other chromosome, including the cassette. With the cassette now on both chromosomes, it is almost certain to be passed on to all offspring.
 The process repeats in the cells of the next generation, with the cassette copying itself so it is on both the paternal and maternal chromosome.
The process repeats in the cells of the next generation, with the cassette copying itself so it is on both the paternal and maternal chromosome.CRISPR gene editing uses a Cas enzyme, which works like a pair of a molecular scissors to cut DNA [3]. In 2014 Kevin Esvelt and his team at MIT outlined a technically feasible way to develop CRISPR-based gene drives[4] and this approach has led to a flurry of research activity and media attention.
The most investigated form of CRISPR-based gene drive is known as a homing gene drive. This works by cutting a target DNA sequence in an animal’s genome and then copying or ‘homing’ itself into the cut site (see box, above). A CRISPR-based homing gene drive is essentially an engineered over-replication gene drive.
To build this system an animal is engineered with a gene drive cassette that encodes the two components of CRISPR: the Cas enzyme and guide RNA. These components, when expressed from one allele, then cut the sister allele on the other (homologous) chromosome. After the sister allele on the homologous chromosome is cut, a cellular process known as homologous recombination results in the gene drive cassette (and any additional genetic payload included in it) being copied onto the homologous chromosome.
This means that both sets of chromosomes have copies of the gene drive cassette – and any animal with two copies of a gene is guaranteed to pass it on to all their offspring. As well as the CRISPR machinery, the gene drive cassette can be engineered to deliver additional DNA sequences with a desired trait. In the offspring generation, the homing process occurs again and will do so in all subsequent generations, resulting in the gene drive spreading through the targeted population[4].
Beyond mosquitoes
To date CRISPR-based homing gene drives have been developed in yeast, fruit flies, two-species of malaria-carrying mosquitoes and most recently in mice[5,6]. Most of the research on CRISPR gene drives thus far has focused on controlling populations of mosquito as vectors of human disease and significant progress has been made towards achieving this goal. Demonstrations of the technology in other species have primarily been proof-of-concept experiments, which have widened the scope of the technology’s potential applications.
Another potential application of gene drives is to control invasive pests. Invasive vertebrate pests, such as cane toads and rabbits in Australia, grey squirrels in the UK, possums in New Zealand and rodents around the globe cause significant damage to the environment, and incur economic and social costs. Control measures include shooting, poisoning, trapping and the release of biological agents, which are largely inadequate and costly, and often lead to unwanted suffering in both target and non-target species.
Gene drives may offer a more cost-effective, humane and species-specific alternative. In most pest species, female procreative capacity is responsible for maintaining the overall population size and so an efficient means of population suppression is to bias the sex ratio in favour of males. A grossly male breeding population will result in a population decline, while an all-male breeding population would lead to eradication.
A homing-based gene drive can achieve this with a gene drive cassette targeted to disrupt the coding sequence of an essential female fertility gene[5]. Males would be unaffected by loss of the female fertility gene, while all females that inherit the drive would be infertile.
 Gene drives could help conservationists spread resistance to CWD in threatened cervids such as moose, although their long generation interval would mean the spread of any resistance genes would take significantly longer than in insect or amphibian models.
Gene drives could help conservationists spread resistance to CWD in threatened cervids such as moose, although their long generation interval would mean the spread of any resistance genes would take significantly longer than in insect or amphibian models. Gene drives could also be used to rescue threatened or endangered animals. For instance chronic wasting disease (CWD) is a form of prion disease in cervids that affects primarily deer, elk and moose. The infectious prions responsible for CWD are caused by misfolding and aggregation of a normal, cellular prion protein (PrPC) and are spread through saliva, urine and faeces. Once an animal is infected, misfolded prion protein builds up in the brain, causing neuronal loss and eventually leading to death. In some states in northern US, prevalence of CWD in deer is up to 35% in free-ranging populations and 90% in captive deer.
Studies in rodents and large animals have shown that animals do not require PrP7 and that gene drives that remove or ‘knock out’ the PrP gene could be used to rescue infected cervid populations from this highly infectious disease. However, the long generation interval in cervids would mean the drive spreads far more slowly through the population than in insect applications, for example.
Rescuing amphibians
Gene drives could also be applied to rescue threatened amphibians from chytridiomycosis. Chytridiomycosis is the disease caused by the fungus Batrachochytrium dendrobatidis, which has been implicated in mass die-offs and the extinction of hundreds of frog species since the 1990s, and threatens up to 50% of amphibians.
As the fungus reproduces mostly asexually, it cannot be controlled with a gene drive itself. However, resistance to B. dendrobatidis infection varies both within and among amphibian species. Major histocompatibility complex (MHC) peptides play an important role in the innate immune system of vertebrates and a specific MHC allele in the lowland leopard frog has been shown to increase survival of infected individuals[8].
Gene drive could be used to spread the lowland leopard frog’s resistant MHC allele through threatened amphibian populations, rescuing species from this killer fungus and potentially saving them from extinction. Careful consideration in the gene drive design would be required to ensure the gene drive does not impair the function of key immune system genes.
In Norway the total cost of sea lice to the aquaculture industry is around £400m per year, and treatments involve chemicals that damage marine ecosystems, affect fish appetite and growth, and mean treated fish can’t be sold for several weeks after treatment. There is also resistance among lice to the three major classes of chemicals used. Self-limiting gene drives could be deployed to locally suppress lice populations in fish farms without affecting the broader marine ecosystem. Another pest species that has significant economic and welfare implications for animal agriculture is the New World screwworm (NWS) fly. NWS causes myiasis, a parasitic infestation of warm-blooded animals as a result of the flies’ larvae growing inside the host and feeding on its flesh. The screwworm is found in all South American countries except Chile and has an overwhelming impact on livestock farming throughout the region. Wildlife is also threatened, especially endangered species. The US is considered NWS-free after a 50-year eradication programme estimated to have cost £1bn. As a potentially quicker and cheaper solution the nations of South America could deploy self-limiting gene drives to locally suppress NWS or a self-perpetuating drive to eradicate the pest from the entire continent, potentially saving wildlife from NWS infestation and increasing agricultural productivity.
Controlling the drive
The main concern with homing gene drive strategies is that these self-perpetuating genetic elements can, in theory, spread indefinitely. There are, however, scenarios and strategies to prevent the duration and extent of this.
First, the spread would be limited by naturally arising resistant alleles that prevent CRISPR from recognising and cutting its target site. These resistant alleles could exist in the population prior to release or evolve when an error in the DNA repair process occurs and a CRISPR-mediated cut is repaired with a new functional DNA sequence (which the CRISPR guide RNA in future generations then does not recognise).
If a gene drive was not stopped by naturally arising resistance, there are ‘stop’ buttons available, theoretically. Gene drives can be inactivated by the release of animals with engineered resistant alleles or even with a ‘reversal gene drive’ that immunises the animal against the original drive.
However, these reactive stop buttons are not ideal. A more proactive approach is needed and scientists are working to develop gene drives that are inherently self-limiting – that is, they only stay in a population transiently and could be localised to targeted populations.
Several self-limiting gene drive concepts have been proposed and it is hoped these systems will soon be realised[9]. One type is a so-called ‘daisy-chain drive’: with this approach the CRISPR components of the gene drive are split up and scattered throughout the genome so that none of them can be copied on its own. Element A can only copy itself if element B is present, and element B can only copy itself if element C is present. And element C, crucially, cannot copy itself at all – it can only be passed on to offspring at the normal rate of 50%.
If daisy-chain animals carrying all three elements were released, all the offspring will inherit element A and B, but only half will inherit element C. In the following generation element B and A will spread, but C will gradually die out. Once it does, B will start to disappear and finally A will too.
Future potential
If robust self-limiting gene drives can be developed, it puts the power in the hands of communities to deploy a gene drive locally, avoiding the need for highly complex geopolitical agreements that will be required for self-perpetuating gene drive systems.
With the realisation of self-limiting gene drives, this technology would be more amenable to applications in animal agriculture and aquaculture, such as the pressing issue of sea lice in fish farming or the eradication of the New World screwworm.
Although gene drive technology still faces significant scientific, political and social hurdles, we are optimistic of its future potential and as such have chosen to highlight prospective applications of the technology in this article. However, it must be noted that before any of these proposed applications are deployed there is a requirement for in-depth analysis of the ecological implications, as well as the need for broad community engagement with those that may be affected by the release of a gene drive.
Each potential application should be considered on a case-by-case basis, but with the rapid progress in this field, it is hoped the risks associated with current homing gene drive strategies are reduced with the realisation of self-limiting gene drive approaches.
For the first time we have the makings of a technology that could allow us to overcome the limitations imposed by conventional genetic inheritance and utilise molecular biology to spread beneficial traits through wild animal populations. Both Mendel and Darwin would certainly be astounded by how far our understanding has come and the benefits gene drives could offer our society and the environment.
Bruce Whitelaw FRSB is deputy director and director of partnerships at the Roslin Institute and professor of animal biotechnology at the Royal (Dick) School of Veterinary Studies. He is currently establishing robust methodology for genome editing in animal systems.
Gus McFarlane AMRSB is a doctoral student at the Roslin Institute focused on developing CRISPR-based gene drive strategies for controlling invasive vertebrate pests. He is interested in the application of genome editing tools to improve the welfare and performance of livestock.
1) Burt, A. & Trivers, R. Genes in conflict: the biology of selfish genetic elements. Harvard University Press (2009).
2) Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proceedings of the Royal Society B: Biological Sciences 270(1518), 921–928 (2003).
3) Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096), 816–821 (2012).
4) Esvelt, K. et al. Concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401 (2014).
5) McFarlane, G. et al. CRISPR-based gene drives for pest control. Trends in Biotechnology 36(2), 130–133 (2018).
6) Grunwald, H. et al. Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566(7742), 105–109 (2019).
7) Richt, J. et al. Production of cattle lacking prion protein. Nature Biotechnology 25(1), 132–138 (2007).
8) Savage, A. & Zamudio, K. MHC genotypes associate with resistance to a frog-killing fungus. Proceedings of the National Academy of Sciences 108(40), 16705–16710 (2011).
9) Min, J. et al. Harnessing gene drive. Journal of Responsible Innovation 5(sup1), S40–S65 (2018).


